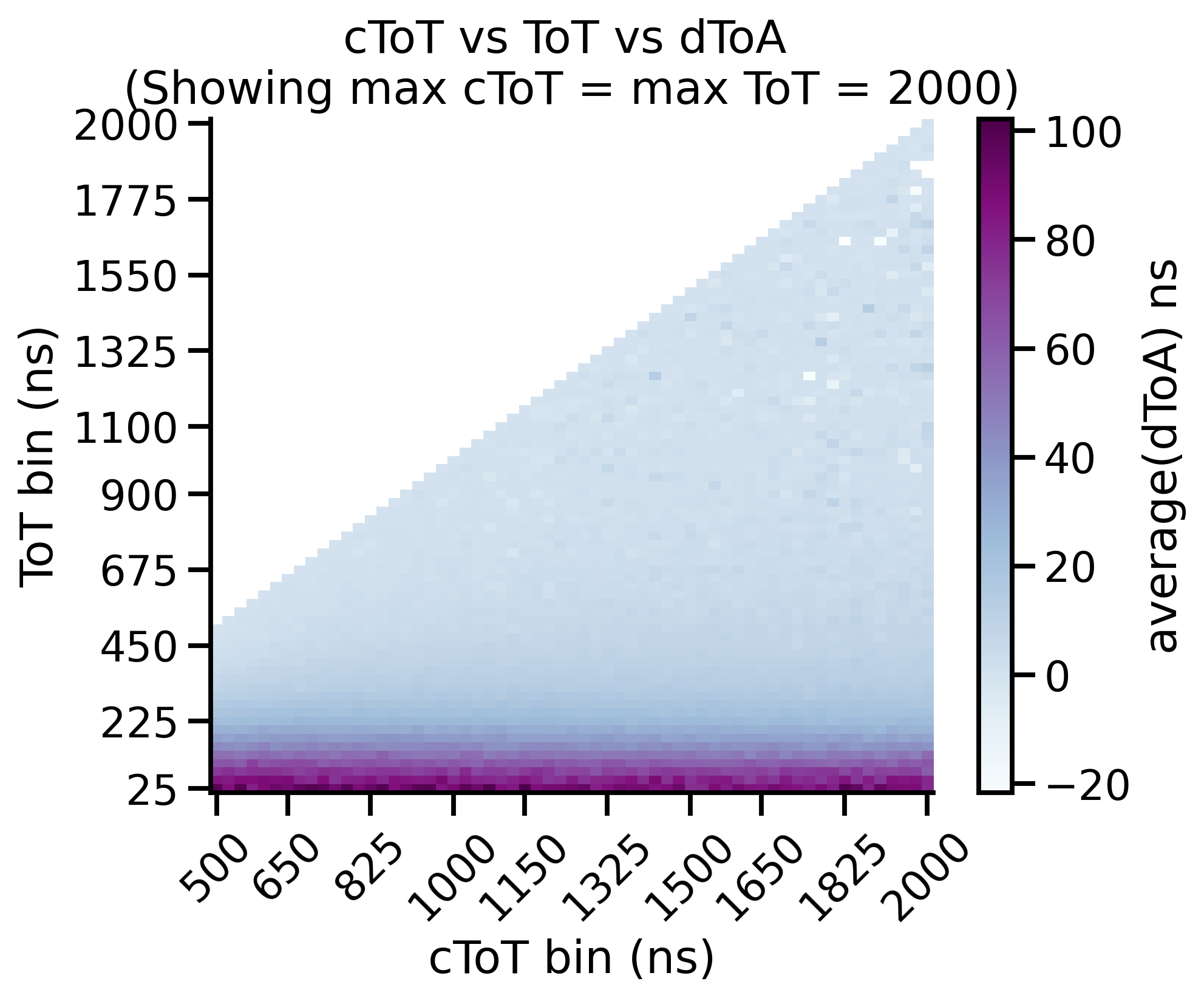
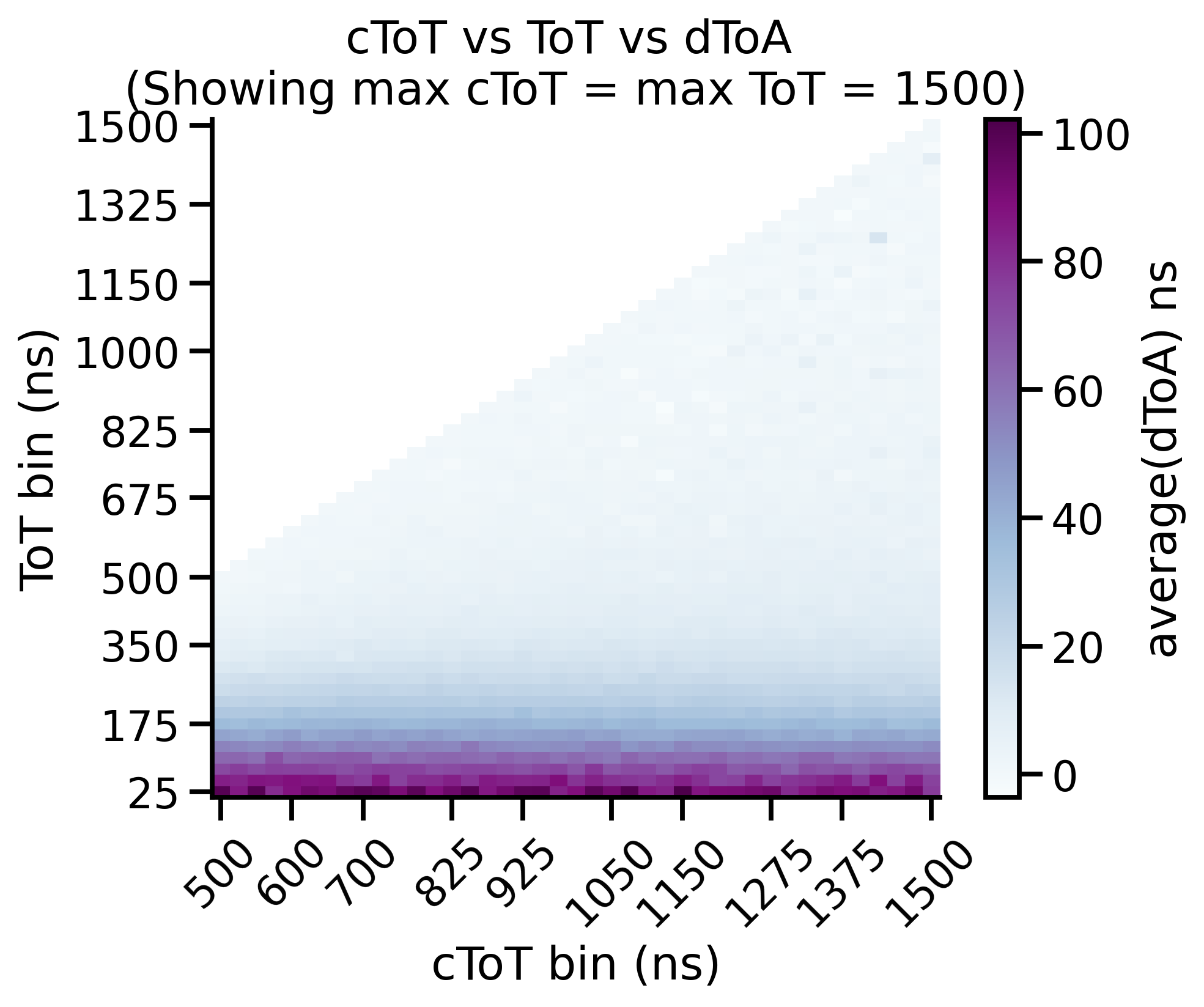
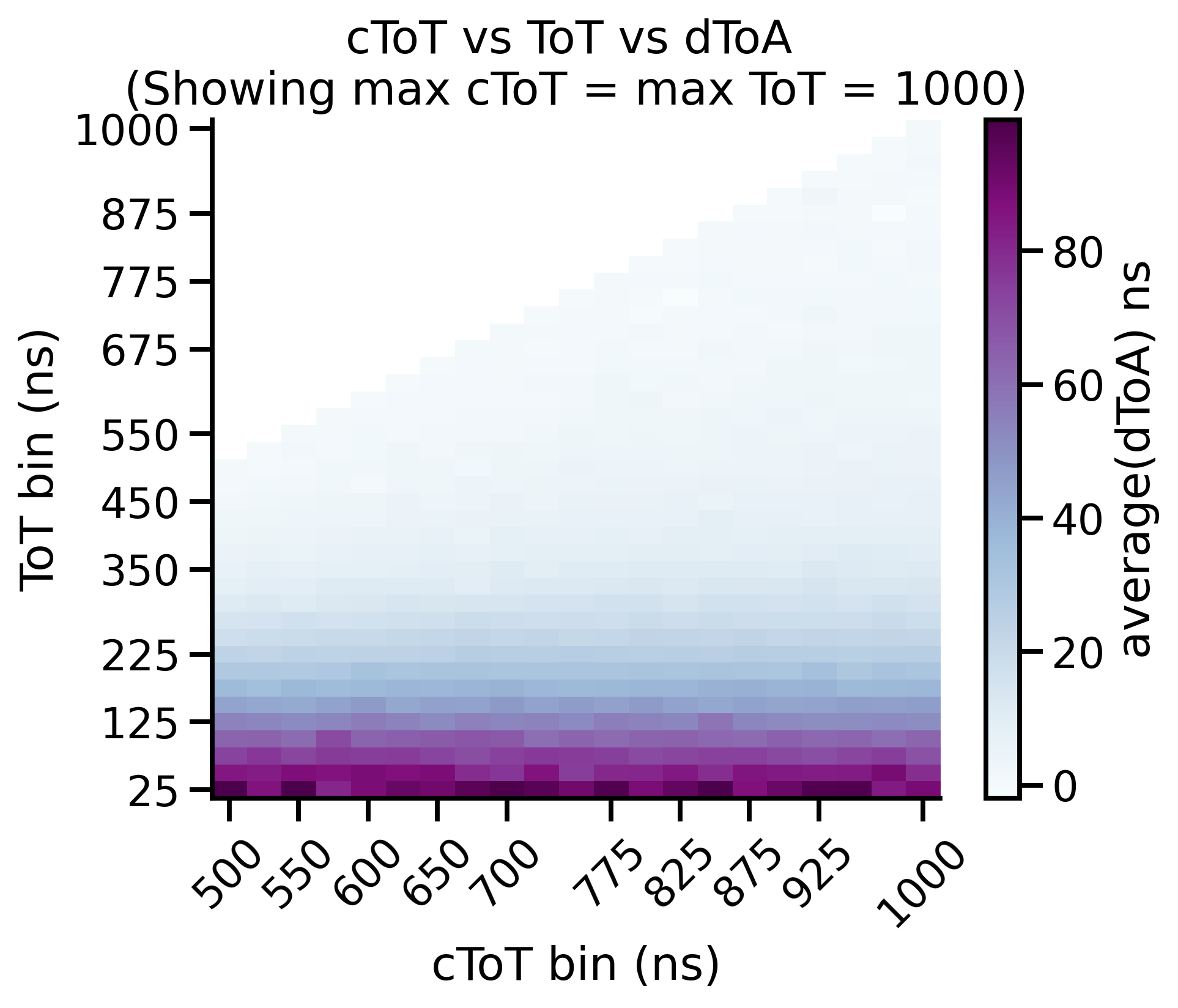
Time-walk Correction: Matrix¶
The timewalk matrix gives important insight into your data. More specifically it shows you where to put the cToT cut parameter. The timewalk lookup table is an average of this matrix, “cut” at a particular cToT point. That is, values lower than this are discarded and values higher are averaged and used to construct the lookup table.
Luna Command¶
tpx3dump process -i /Users/Ciaran/atlassian-bitbucket-pipelines-runner/temp/e71169e4-520a-5b30-a5ab-ee8a44eb5fac/build/docs/source/_static/example_data.tpx3 -o /Users/Ciaran/atlassian-bitbucket-pipelines-runner/temp/e71169e4-520a-5b30-a5ab-ee8a44eb5fac/build/docs/source/_static/example_data.hdf5 --eps-t 150ns --eps-s 1 --ctot-cut 500
Python Script¶
Plot the timewalk matrix¶
1import os, sys
2from typing import *
3import lmfit
4import pandas as pd # ensure you have `pip install pandas`
5import warnings
6import seaborn as sns
7import matplotlib.pyplot as plt
8from matplotlib import cm
9from matplotlib.colors import Normalize
10import numpy as np
11
12
13warnings.filterwarnings("ignore") # suppress warnings from plotting libraries.
14
15sns.set_context(context="talk")
16
17# add some paths to PYTHONPATH
18for directory in ["..", "."]:
19 sys.path.append(os.path.abspath(os.path.join(os.path.dirname(__file__), directory)))
20
21# on our system "EXAMPLE_DATA_HDF5" refers to the absolute path
22# to a hdf5 file generated by luna. Replace with your own!
23from env_vars_for_docs_examples import EXAMPLE_DATA_HDF5, PLOTS_DIRECTORY
24
25# re-use functions from previous example
26from ex2_read_data_time_units import load_timewalk_matrix, TimeUnit
27
28
29def plot_timewalk_matrix_heatmap(timewalk_matrix: pd.DataFrame, max_ctot_and_tot_to_show: Optional[List[int]] = None):
30 """
31 Plots a heatmap of the timewalk matrix.
32
33 This function generates and saves heatmaps of the timewalk matrix, showing the relationship between
34 CToT (Corrected Time over Threshold) and ToT (Time over Threshold) bins, and their corresponding
35 AverageDToA (Average Delta Time of Arrival) values. The heatmaps are saved as PNG files.
36
37 Parameters:
38 -----------
39 timewalk_matrix : pd.DataFrame
40 A DataFrame containing the timewalk matrix data. The DataFrame should have the following columns:
41 - 'CToT': Corrected Time over Threshold
42 - 'ToT': Time over Threshold
43 - 'AverageDToA': Average Delta Time of Arrival
44 - Other columns may include 'SumSquareDiff', 'Count', 'Std', 'Sem', etc.
45
46 max_ctot_and_tot_to_show : Optional[List[int]], default=None
47 A list of integers specifying the maximum CToT and ToT values to show in the heatmap. If None,
48 the default value [1024 * 25] is used. Each value in the list will generate a separate heatmap.
49
50 Returns:
51 --------
52 None
53 The function does not return any value. It saves the generated heatmaps as PNG files in the
54 specified directory.
55
56 Raises:
57 -------
58 AssertionError
59 If `max_ctot_and_tot_to_show` is not a list of integers.
60
61 """
62 if max_ctot_and_tot_to_show is None:
63 max_ctot_and_tot_to_show = [1024 * 25]
64
65 for max_tot in max_ctot_and_tot_to_show:
66 assert isinstance(max_ctot_and_tot_to_show, list), "max_ctot_and_tot_to_show should be a list of integers"
67
68 timewalk_matrix_avg = timewalk_matrix.pivot(index="ToT", columns="CToT", values="AverageDToA")
69 print(timewalk_matrix_avg.head())
70 data = timewalk_matrix_avg.loc[timewalk_matrix_avg.index <= max_tot, timewalk_matrix_avg.columns <= max_tot]
71
72 fig, ax = plt.subplots()
73 sns.despine(fig=fig)
74 img = ax.imshow(data.values,
75 origin="lower",
76 cmap="BuPu",
77 aspect="auto",
78 )
79 ax.set_ylabel("ToT bin (ns)")
80 ax.set_xlabel("cToT bin (ns)")
81
82 xticks = np.linspace(0, data.shape[1] - 1, num=min(10, data.shape[1]), dtype=int)
83 yticks = np.linspace(0, data.shape[0] - 1, num=min(10, data.shape[0]), dtype=int)
84
85 ax.set_xticks(xticks)
86 ax.set_yticks(yticks)
87 ax.set_xticklabels(data.columns[xticks], rotation=45)
88 ax.set_yticklabels(data.index[yticks])
89
90 c = plt.colorbar(img)
91 c.set_label('average(dToA) ns')
92 plt.title(f"cToT vs ToT vs dToA \n(Showing max cToT = max ToT = {max_tot})")
93 fname = os.path.join(PLOTS_DIRECTORY, f"ex8_timewalk_matrix_heatmap_{max_tot}.png")
94 plt.savefig(fname, bbox_inches='tight', dpi=300)
95
96
97
98if __name__ == "__main__":
99 toa_unit = TimeUnit.Nanoseconds
100
101 timewalk_matrix = load_timewalk_matrix(EXAMPLE_DATA_HDF5, toa_unit=toa_unit)
102 print("Timewalk Matrix:")
103 print(timewalk_matrix.head().to_string())
104 if timewalk_matrix is not None:
105 tot_thresholds = [ 500, 1000, 1500, 2000]
106 plot_timewalk_matrix_heatmap(timewalk_matrix, tot_thresholds)
Script Output¶
Example Output¶
<KeysViewHDF5 ['Clusters', 'ExposureTimeBoundaries', 'PixelHits', 'TimewalkLookupTable', 'TimewalkMatrix']>
Timewalk Matrix:
CToT ToT AverageDToA SumSquareDiff Count Std Sem
0 500 25 99.356621 94192336.0 17 23.538746 5.708984
1 500 50 84.650749 80431408.0 17 21.751457 5.275504
2 500 75 73.153412 76722296.0 44 13.204880 1.990710
3 500 100 63.130474 104241200.0 57 13.523288 1.791203
4 500 125 54.665501 144652992.0 71 14.273634 1.693969
CToT 500 525 550 ... 6675 6825 6850
ToT ...
25 99.356621 85.677086 99.588829 ... NaN 23.437500 20.833334
50 84.650749 83.046875 86.979172 ... 14.062500 18.750000 20.312500
75 73.153412 76.286766 72.593735 ... 14.062500 16.406252 13.541668
100 63.130474 63.586956 60.998768 ... 13.020833 7.812500 8.593750
125 54.665501 53.568096 51.769302 ... 12.500001 NaN 11.328125
[5 rows x 189 columns]
CToT 500 525 550 ... 6675 6825 6850
ToT ...
25 99.356621 85.677086 99.588829 ... NaN 23.437500 20.833334
50 84.650749 83.046875 86.979172 ... 14.062500 18.750000 20.312500
75 73.153412 76.286766 72.593735 ... 14.062500 16.406252 13.541668
100 63.130474 63.586956 60.998768 ... 13.020833 7.812500 8.593750
125 54.665501 53.568096 51.769302 ... 12.500001 NaN 11.328125
[5 rows x 189 columns]
CToT 500 525 550 ... 6675 6825 6850
ToT ...
25 99.356621 85.677086 99.588829 ... NaN 23.437500 20.833334
50 84.650749 83.046875 86.979172 ... 14.062500 18.750000 20.312500
75 73.153412 76.286766 72.593735 ... 14.062500 16.406252 13.541668
100 63.130474 63.586956 60.998768 ... 13.020833 7.812500 8.593750
125 54.665501 53.568096 51.769302 ... 12.500001 NaN 11.328125
[5 rows x 189 columns]
CToT 500 525 550 ... 6675 6825 6850
ToT ...
25 99.356621 85.677086 99.588829 ... NaN 23.437500 20.833334
50 84.650749 83.046875 86.979172 ... 14.062500 18.750000 20.312500
75 73.153412 76.286766 72.593735 ... 14.062500 16.406252 13.541668
100 63.130474 63.586956 60.998768 ... 13.020833 7.812500 8.593750
125 54.665501 53.568096 51.769302 ... 12.500001 NaN 11.328125
[5 rows x 189 columns]